microbes in animal landscapes
Welcome to the website of:

Mapping the impact of microbiota on infection outcomes.
About me.
Welcome to my homepage! I am a microbiologist striving to better understand general mechanisms how our microbes interact with us and our health. Therefore, I view the human body from the perspective of microbes – a vast and diverse ecosystem. My passion for discovering general principles in natural chaos drives me, aligning with the ultimate goal of developing innovative therapies to improve health and well-being.
Current address: Helmholtz Centre for Infection Research, Bacterial Infection Ecology, Inhoffenstrasse 7, 38124, Braunschweig
Projects.
Spatial Ecology of Pathogen Resistance (SpatialGut)
There exist dense microbial communities within us that act as a protective barrier against pathogens. However, a notable gap in our understanding is to how these communities are organised in the intestine and how this organisation links to health outcomes.
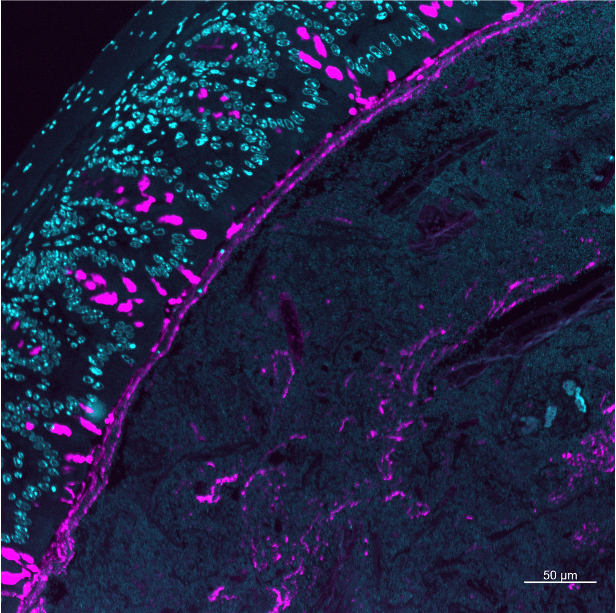
This project aims to generate a comprehensive single cell map of gut microbiota in the context of infection resistance. This enables us to address fundamental questions on how the spatial organisation drives infection resistance. Therefore, I iteratively map and manipulate the spatial microbiome organisation to assay its impact on colonisation resistance against the pathogen model Salmonella enterica.
We establish HG23, a new humanised mouse model, that allows now to map entire microbiota at the single cell level in relation to the host and invading pathogens.
Ultimately, this provides new basic insights guiding the rational design of beneficial microbial communities to improve infection resistance.
Previous PROJECTS.
Virus-host-microbe-immune interactions in sponges (SpoVir)
This work establishes the integral role of viral communities in marine sponges – the oldest known animal-microbe symbiosis. One of the key findings of the research was the identification of the phage protein ANKp, which helps symbiont bacteria evade the eukaryotic immune system. This was the first time a phage with such an immunomodulatory effect had been recognized. The study’s results have significant implications for the fields of immunology, ecology, and symbiosis research, particularly towards phage derived immune therapy.
The study’s findings also revealed that despite filtering large amounts of seawater, marine sponges possess unique viral signatures and replication strategies that are distinct from other environments. This emphasizes the importance of marine animals as a repository for novel viral diversity. The study’s results contribute to a deeper understanding of the mechanisms of phage interaction with the sponge’s early immune system, providing insight into the potential application of phage therapy in treating various illnesses.
This work was covered in The Scientist by Catherine Offord (here) and featured by Brittany A. Leigh (here)
Integrative symbiont physiology (iSymbP)
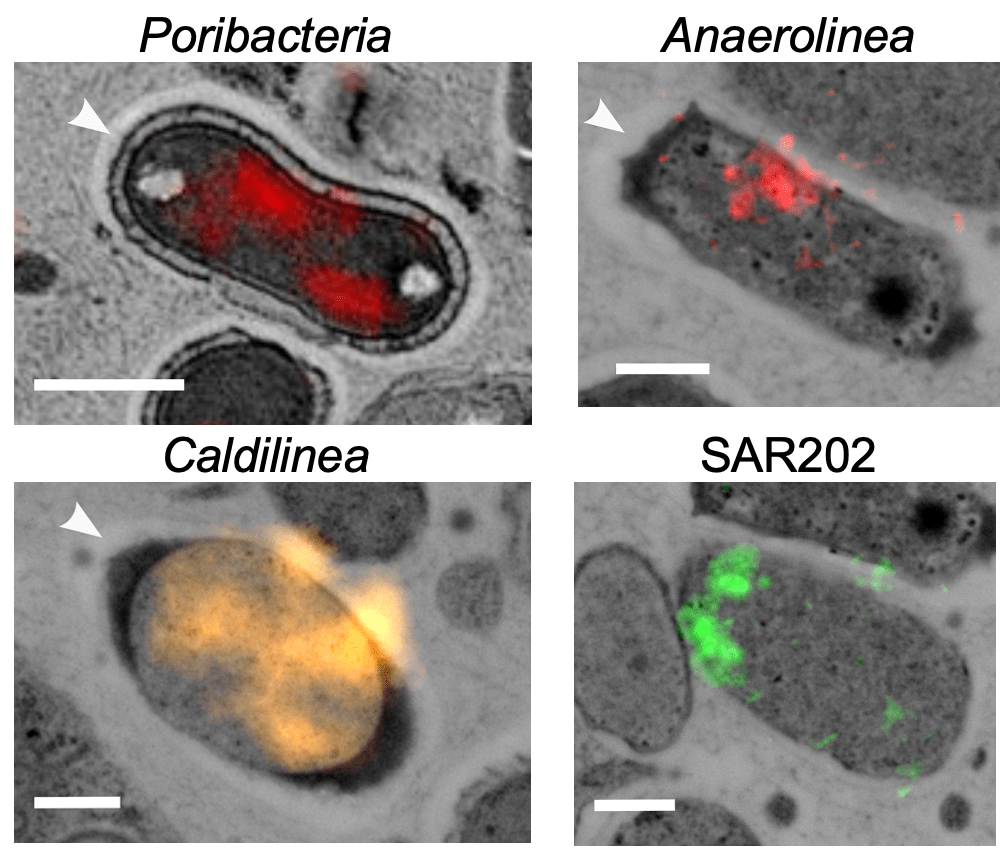
The majority of symbionts in nature remain uncultured, making it difficult to understand their physiology and impact on animal hosts. This project addressed this challenge by developing new methods to study uncultured sponge symbionts. The study focused on Poribacteria, a cosmopolitan symbiont of sponges that has evaded cultivation so far. Genome-centered metatranscriptomics was used to analyze gene expression, while FISH-CLEM imaging enabled simultaneous identification of specific viruses, symbionts, and transcripts in the microbiome, providing new insights into the morphology and physiology of the symbiont. Specifically, The FISH-CLEM imaging method identified structural adaptations of Poribacteria to store and process animal-derived compounds. This major new imaging method allows for the simultaneous identification of specific microbes, cellular structures, and target proteins at high resolution in their environmental context.
Collaborations & Further Works
If you are interested in viewing a comprehensive list of my projects and collaborations, I invite you to visit my Google Scholar page, which includes information on further research topics. Some examples of my work include establishing a high throughput method for isolating marine micro-algae (Jahn et al 2014), exploring Borrelia landscape ecology (Millins et al 2018; see picture), and collaborating on projects related to gut immune pathology (Stengel et el 2020) and infection (Hildebrand et al 2019). Thank you for your interest in my research.

If you have collaboration ideas, want to discuss science, or want to say hi, please get in touch.
TEACHING.

As someone who has benefited greatly from passionate teachers and mentors, I am driven to inspire the next generation in the same way. My philosophy is that there are no bad questions, and that we must fight for inclusive environments of trust that encourage curiosity and inquiry – essential for fostering productive and efficient learning outcomes. Further, I belief it is important not only to teach what is already known, but also to explore the unknown and challenge assumptions. This approach not only deepens students’ understanding, but also encourages critical thinking and creativity.

Microbiome Class (left), Practical (right), Oxford
Stages in Pictures.
I am grateful for the perspective I have gained through the opportunities to study host-microbe interaction across a diverse range of ecosystems, from the Arctic to the Mediterranean, and from land to sea. By exploring the earliest animal-microbe interactions in sponges to those in the complex organisms committed to advancing our understanding of these microbial worlds.

Spatial Epidemiology, Scotland
Sampling 1200 ticks for Borrelia

Throughput isolation of arctic algae, Norwich, England
Fast and easy methods to transform environmental samples to characterised pure cultures.
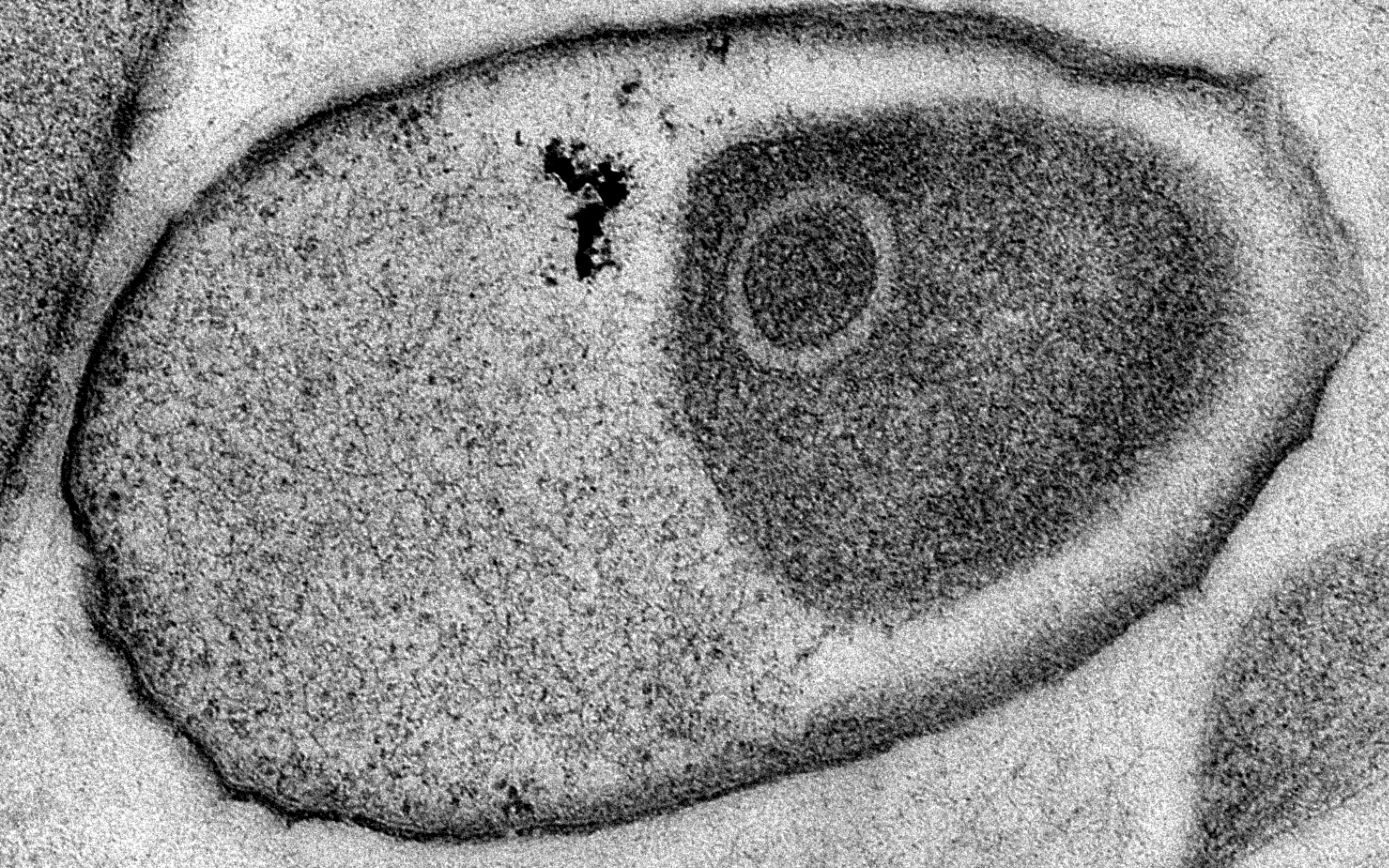
Sponge Symbiont in host matrix, GER
Highly sub-structured symbiont of unknown identity residing between collagen fibrils.

Sponge viromics, North Catalonia, Spain
Sampling sponge associated-microbiota.
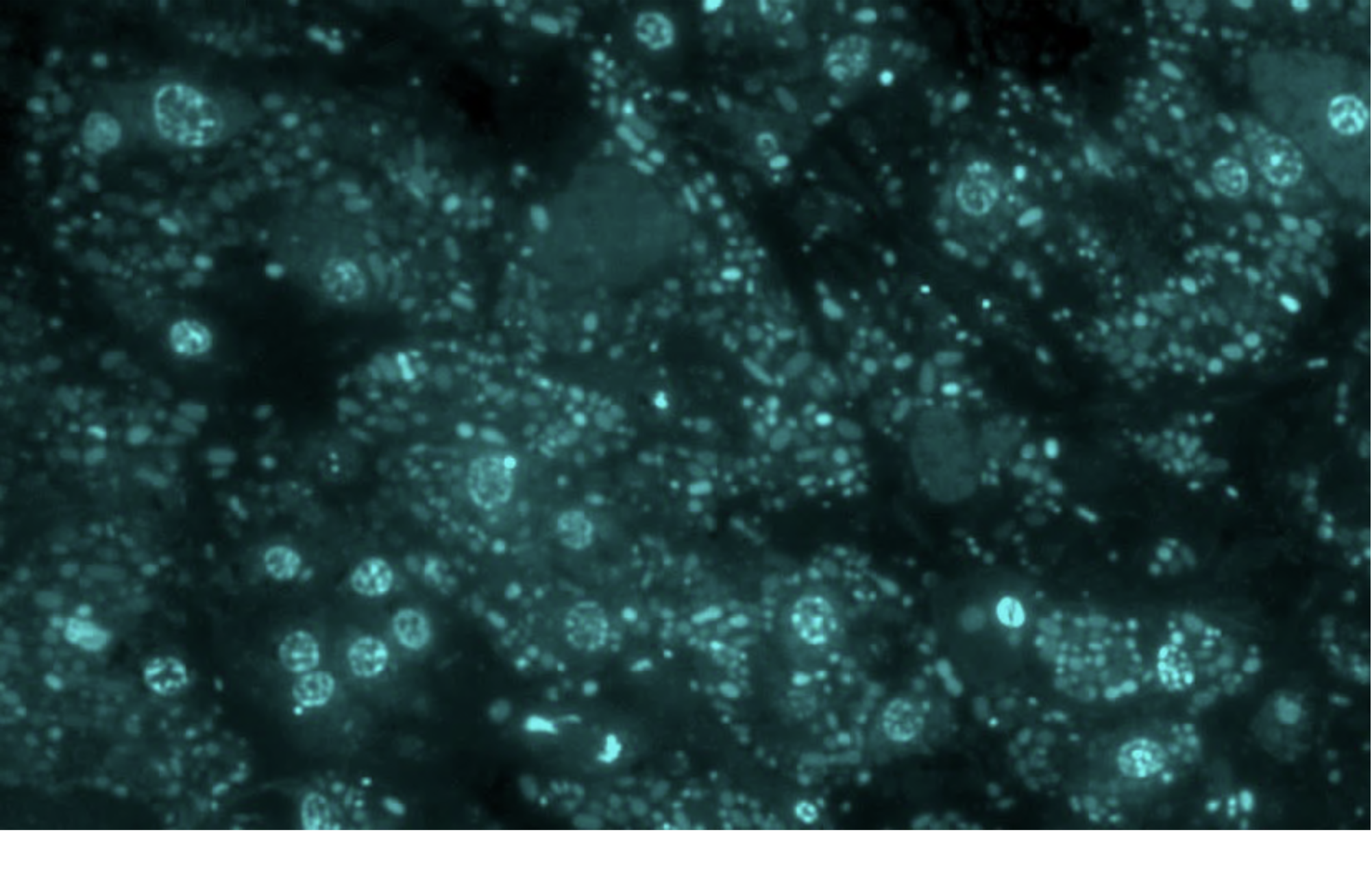
Human Gut Microbiota, GER
The first image I captured from a human patient’s faeces, which inspired me to shift my focus from capturing symbioses across landscapes to exploring the intricate and diverse landscapes that exist within us.
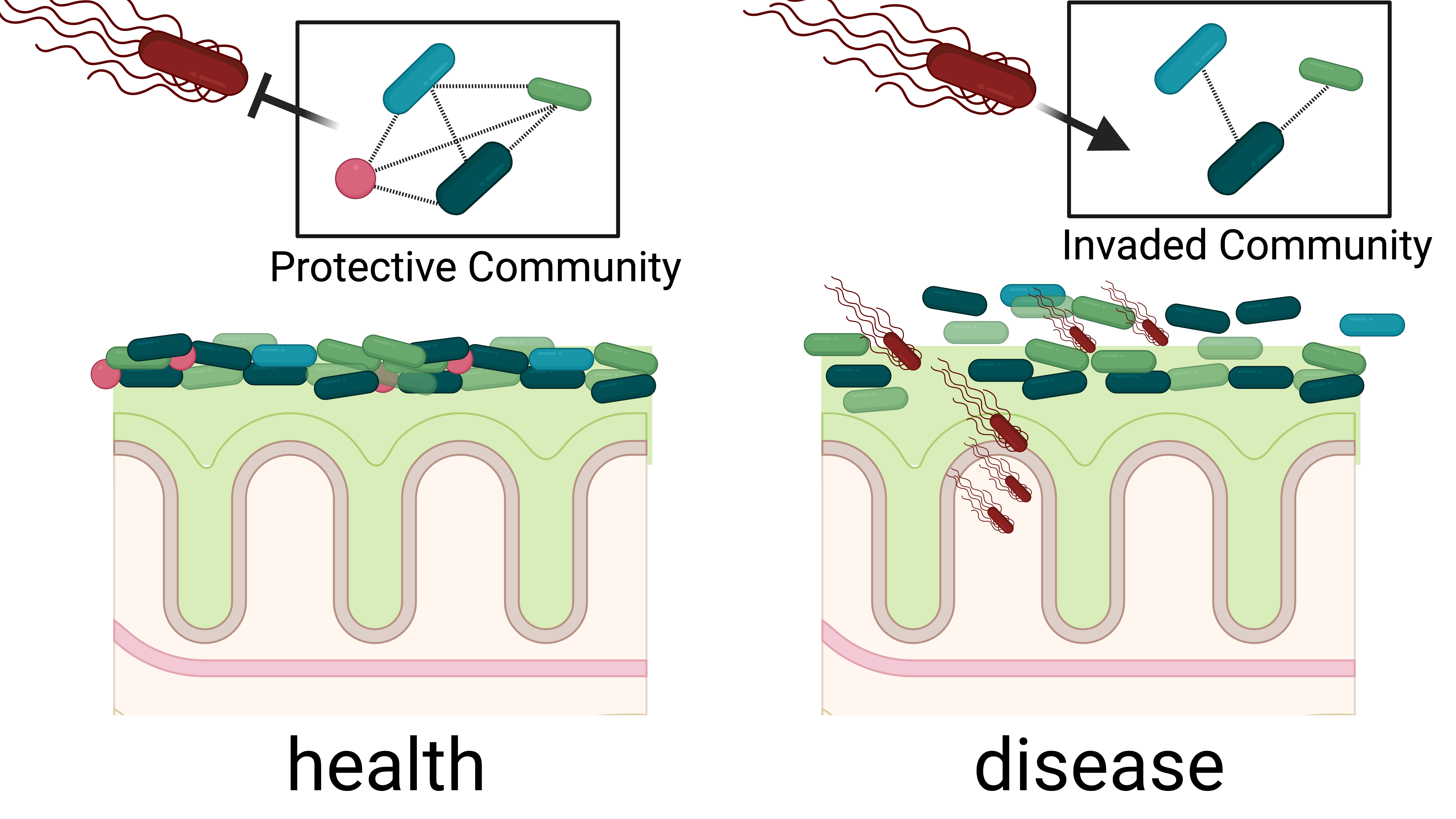
Spatial Infection Ecology of gut Microbiota, Oxford, UK
Models to study microbial spatial ecology within us in inflammation & infection.